Yesterday, I explain a method to download nr database for a particular organism. In this post let us discuss another approach to download non redundant sequence of a given organisms form NCBI. Let me clear that I love the first method because of its simplicity. Steps of this method are as follows
Also readPart- I : How to Download nr Database For a Particular Organism
- Go to the NCBI home page HERE.
2. Choose PROTEIN as target database and Search with all [filter] query
3. On the result page, you will get the list of organism and the number of sequences of that organism in bracket.
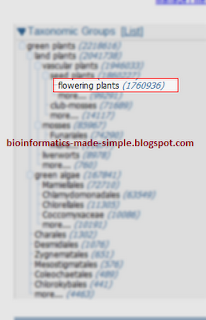
4. Click on the list/tree to change the view.
5. Choose the taxonomic group to which your organism belong to. (For example, flowering plant if I am looking for Arabidopsis)
6. You can narrow down the classes
7. Finally You can export you sequences as I explain in step 6 of my previous tutorial.
I know that there may be so many other easier method to download nr database for a specific organism. If you know anyone, please do share with us





Post a Comment